|
|
Genetic Management of the Mexican Wolf Captive Population: By Philip S. Miller
Introduction Although persisting as a subject of often intense debate within many realms of the conservation community, the management of animal populations in captivity remains an important and often vital component of an endangered species management strategy. The final goal of most captive management programs includes the reintroduction of healthy captive-born individuals to wild habitats that best suit their biological needs and are as free as possible from human interference. To help achieve this goal, zoo population managers work hard to secure the health of these individuals – not only in terms of their physical well-being, but also with regards to their very genetic make-up. Specifically, we want these animals to be as similar as possible to their wild-born brethren: to be free of genes that may “creep” into the population from accidental breeding with closely-related species, and to be the product of matings between healthy, unrelated animals so that concerns about the detrimental effects of inbreeding (pairing related individuals) can be reduced or, hopefully, eliminated completely. For our programs to be successful, careful genetic management of these populations – using high-tech scientific approaches ranging from detailed DNA laboratory studies to sophisticated computer-based statistical analysis of genetic data – is a necessity. The conservation history of the Mexican gray wolf (Canis lupus baileyi) provides an excellent example of the successful use of scientific analysis in captive population management, as well as the practical difficulties of conserving such critically endangered species. The original captive population dates back to the late 1970s when six wolves were captured from Chihuahua and Durango in Mexico and placed in zoos in the United States. A decade later, only four of these original founders – just three males and a lone female – had offspring in the captive population. Since its inception, the population that is now known as the McBride lineage has been carefully managed using the AZA’s Species Survival Plan (SSP) methodology for the maximum retention of the original genetic variation collected from the wild in these four animals. Soon after the original founders were brought into captivity, wolf biologists realized that the capture of additional animals was not going to be possible. This was going to have serious consequences for the long-term viability of the McBride lineage since, with only one female founder, all the descendent wolves would be related as (at least) half-siblings and rates of inbreeding would quickly begin to increase. However, zoo population managers had an opportunity to reverse this trend with an option that rarely presents itself to those managing endangered species in captivity. It turned out that two other populations of Mexican wolves existed in captivity: the Ghost Ranch lineage in New Mexico and the Aragon lineage in Mexico City. While each of these lineages were themselves founded by a very small number of individuals (most likely a single pair each), and therefore became highly inbred in a short period of time, it would be possible to dramatically reduce both the frequency of inbreeding and its potential consequences in the McBride lineage by integrating the two additional lineages into the SSP population. The science of population genetics tells us that, even if two individuals are themselves highly inbred but unrelated to each other, their offspring will not be inbred and, most likely, more healthy than their inbred counterparts. The future of the captive population was looking brighter and brighter. But the situation was far from simple. There were many questions about the genetic status of all three lineages that had to be addressed by scientists around the country before wolves from different bloodlines could be paired together. Specifically, geneticists were faced with the following unknowns:
Sophisticated molecular genetics laboratory techniques were used to collect the required data, and a variety of statistical analysis methods were then employed to provide guidance in answering these questions. A Molecular Genetic Evaluation of the Three Mexican Wolf Lineages A variety of different analytical techniques were used to assess the genetic purity of the McBride, Ghost Ranch and Aragon lineages of Mexican wolves. Simple morphological analysis of skulls from Mexican wolves, coyotes and dogs suggested that the three wolf lineages were indeed pure, but the results were not definitive. It turns out that nutrition and other aspects of the captive environment can, over time, lead to higher levels of variation in skeletal morphology compared to what would be seen in more “wild” samples of northern gray wolves, coyotes, or domestic dogs. More sophisticated techniques of analysis, that would perhaps be less influenced by the physical nature of the captive environment, would have to be used to shed more light on this issue. The assessment of molecular genetic variation has been used for many years to determine the extent of divergence both within and between different populations (samples) of wildlife. Once specific genes are identified across the genome of the species in question, scientists are able to identify how many different versions of that gene exist among individuals in a given population. Different versions of a given gene are called alleles. The identification of different alleles at a genetic locus, as well as their number and similarity in DNA sequence, can be analyzed to determine the degree of relatedness of animals within and between populations. The greater the number of alleles that are shared across two populations, the more related they are. Using this technique, we expected to be able to find alleles that are unique to and therefore diagnostic of domestic dogs, coyotes, gray wolves, or Mexican wolves. Once this “library” of unique alleles is built for each type of canid, specific animals can be tested to see if they have any one of these diagnostic alleles. If we find that a Mexican wolf has a gene that is typically diagnostic of domestic dog, we can conclude that this individual is likely to be the direct descendant of a hybrid mating between these two very different lineages. Our colleagues at the University of California, Los Angeles and elsewhere developed a detailed “library” of genetic variation across a broad diversity of North American canids. Each “book” in this “library” contained detailed information on the different types of alleles found in different populations of coyotes, northern gray wolves, and Mexican wolves, as well as different breeds of domestic dog. In some cases, these alleles were further characterized by an analysis of their complete DNA sequences, thereby allowing for an even more fine-scale analysis of genetic differences between individuals and populations. In particular, the genes we were studying are known as microsatellites – genes that are very similar to those used in the popular method of DNA fingerprinting that is often used in courtroom trials of suspected criminals. Microsatellite genes are highly variable – that is, there are many different types of alleles for any one given gene – and the differences between these alleles are often quite small because the different alleles change their sequence through mutation quite rapidly. These characteristics make microsatellites the genes of choice to use in identifying differences between closely-related species or populations. We then began to study this “library” to identify similarities and differences in the alleles across the different species. Usually, scientists must turn to complex methods of mathematical analysis to tease apart the many subtle differences between DNA sequences so that a more definitive conclusion about levels of similarity can be made. Our job here was no exception.
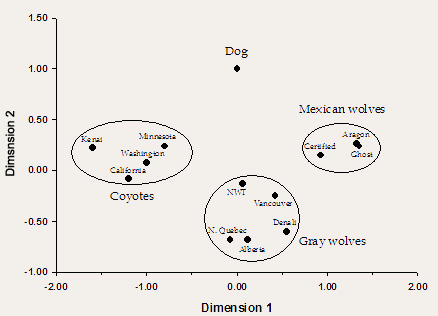
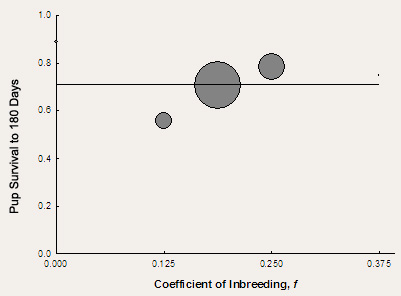
Figure 1. Statistical analysis of microsatellite allele frequency data among We turned to the scary-sounding method of multidimensional scaling to study the differences among our canid populations. In its simplest form, multidimensional scaling is a type of statistical analysis that can describe nearly any measure of similarity between a set of objects as physical distance between them. Therefore, the more similar two objects are to each other, the closer together they cluster in two-dimensional space. In our analysis, the measure of similarity was the frequency of different alleles that had been found at 20 different microsatellite genes in coyotes, dogs, northern gray wolves, and Mexican wolves. The results of our analysis are shown in Figure 1 (don’t worry about the scales on either the X-axis or Y-axis; for our purposes they simply define a basic measure of arbitrary distance between points). Using this approach, we see that the Mexican wolf lineages cluster together to the right, away from the other taxa, indicating that they share the closest common ancestry and are distinct from gray wolves and, more importantly for our purposes, coyotes and dogs. Because of this distinct clustering, we can begin to feel more confident in concluding that our Mexican wolf lineages have little to no genetic signature of hybridization with dogs or coyotes. A more detailed analysis of the presence of different microsatellite alleles revealed that there was one unique dog and one unique coyote allele present in any of the three Mexican wolf lineages. However, each of these alleles was found in very low frequency in Mexican wolves and were likely introduced into the McBride lineage by single individuals that had no other morphological or genetic evidence of ancestry to non-wolf taxa. Importantly, we found no evidence of ancestry to dogs or coyotes in either the Ghost Ranch of Aragon lineages – those for which specific concerns about their ancestry had been expressed. Taken together, these analyses allowed us to conclude that each of the three Mexican wolf lineages were pure wolf – free from hybridization with coyotes or domestic dogs. Plans for the integration of these three lineages could move ahead with confidence. Our same genetic “library” could also be used to determine if two of the original wild-caught Mexican wolves once thought to be unrelated founders of the McBride captive lineage (identified by studbook numbers male 2 and female 5) were in fact related. Using methods akin to DNA fingerprinting, we can determine how similar their microsatellite “fingerprints” are and then make a judgment about their extent of relatedness. Unfortunately, it is nearly impossible to be 100% certain about their degree of relatedness using this type of analysis; there is simply not enough information to be able to make an unambiguous statement about the exact level of kinship between two individuals. However, there are enough data that we can make statements about the probability that two individuals are or are not related. Using our microsatellite data, we found that there was a relatively high degree of genetic similarity between founder 2 and founder 5 – high enough that we could not say that these two individuals were unrelated. In fact, the degree of similarity was consistent with a hypothesis that male 2 was the son of female 5 and the brother of other individuals known to be the offspring of this same female. These findings suggest that there were only three founders in the captive population, and not four as was originally assumed. To assume that there were four founders when the evidence points to only three could further jeopardize the future viability of the captive population by breeding individuals that we now believe to be related. Therefore, the Mexican Wolf SSP officially declared the existence of three founders for the McBride lineage in the mid-1990s. Inbreeding Depression in the McBride Lineage With a reduction in the number of founders of the McBride lineage from four to three, a number of matings once thought to be between unrelated individuals are now known to be inbred matings between mother and son. In other words, the frequency of inbreeding rose dramatically in revised genetics calculations once the new pedigree structure was adopted. This finding made more urgent the need to evaluate the impacts of inbreeding on the health of the McBride lineage. Once again, statistical analyses are available to help us study this phenomenon. Careful studbook records have been kept for this lineage since its inception, so we know the parents and life history of each pup born into captivity. With this information, we can calculate the coefficient of inbreeding for each individual. This coefficient is 0.0 in the offspring of completely unrelated parents and increases to a theoretical maximum of 1.0 as the parents become more related (e.g., offspring of matings between siblings are more inbred than the offspring of matings between cousins). Studies of many different captive mammal populations suggest that the survival of juveniles (i.e., pups) declines as their inbreeding coefficient increases. The studbook data – which includes the date of death of each individual as well as its birthdate and its parents – allows us to make a similar assessment for the McBride lineage. The results of our analysis are presented in Figure 2. The straight line in the graph shows the best fit to the full dataset using a statistical technique called regression analysis and, therefore, shows most clearly the relationship between the degree of inbreeding in newborn Mexican wolves and their survival to 180 days. This line is essentially flat and horizontal, indicating that there is effectively no difference in survival between non-inbred wolves (f = 0.0) and those that are relatively highly inbred (f = 0.375). We concluded from this analysis that there is no evidence for inbreeding depression affecting pup survival in the McBride lineage of Mexican wolves. A similar type of analysis also showed no evidence for an inbreeding effect in litter size across females in the lineage. However, a more recent analysis has demonstrated the potential for inbreeding depression for body size in captive Mexican wolves, but it is too early to tell if this phenomenon is either statistically sound or biologically important.
Figure 2. Pup survival in the McBride lineage of Mexican wolves as a Although we have discussed in the scientific literature a series of limitations to these analysis – both biological and statistical –we remain confident that the potential for inbreeding depression across a suite of important traits affecting overall viability of Mexican wolves is low. This has important consequences for our more general assessment of the captive population’s long-term viability, and makes the integration of this lineage with Ghost Ranch and Aragon an even more attractive management strategy. Conclusion Conservation geneticists have brought a number of sophisticated analytical tools to bear on questions surrounding the genetic constitution of the captive population of Mexican wolves in North America. Using these methods, we have concluded that the three lineages which collectively make up the North American captive wolf population show no demonstrative sign of hybridization with local coyote or domestic dog populations. Because of this analysis, and the subsequent mixing of the lineages as dictated by continued genetic monitoring, the number of founders in the captive population has now increased from just three to seven – meaning that the estimated total amount of genetic variation sampled from the original wild population of wolves has now also increased from 83% to almost 93%. While the expected levels of genetic diversity retained within this population over time fall below those that typically characterize an ideally healthy captive population, the increase in founder gene diversity through this mixing of lineages has nevertheless been a major improvement to the prospects for the population’s future. Perhaps of significantly greater benefit to the management of this population is the dramatic reduction of inbreeding brought about by the mixing of lineages. A broad diversity of mating options could now be identified that would not measurably compromise the genetic integrity of the population, and it was even thought that aspects of population performance such as average litter size would increase in the presence of this new infusion of genetic material into the population. At least in part, these expectations appear to be reality at the present time, although more detailed analysis with larger sample sizes will be necessary in the years ahead to make a more definitive conclusion. All in all, the management of the Mexican wolf in captivity is an outstanding example of the collaborative use of sophisticated scientific principles and tools in the conservation of endangered species. When an effort like this is combined with practical, participatory management across nearly 50 zoological institutions that house the species, the future of the Mexican wolf looks better with the birth of each pup. Further Reading Fredrickson, R., and P.W. Hedrick. 2002. Body size in endangered Mexican wolves: Effects of inbreeding and cross-lineage matings. Animal Conservation 5:39-43. Hedrick, P.W., P.S. Miller, E. Greffen, and R.Wayne. 1997. Genetic evaluation of the three captive Mexican wolf lineages. Zoo Biology 16:47-69. Kalinowski, S.T., P.W. Hedrick, and P.S. Miller. 1999. No inbreeding depression observed in Mexican and red wolf captive breeding programs. Conservation Biology 13:1371-1377. |
|